Identification of key genes and pathways in obstructive coronary artery disease by integrated analysis
Introduction
Obstructive coronary artery disease (OCAD) is included in coronary artery disease, defined as one or more blocked coronary arteries ≥50% through quantitative detection of coronary angiography, commonly falling into coronary arterial stenosis of stage 3–4, which the pathological basis is mainly coronary atherosclerosis. The main clinical symptoms are palpitation, chest pain, chest tightness, dyspnea and so on, relevance high risk of myocardial infarction, heart failure, pulmonary embolism, congestion and edema of the lungs, or cardiac and respiratory arrest (1,2). OCAD is the main reason of incidence rate and death toll worldwide, with a prevalence ranging from 75% to 95%, far more than non-obstructive coronary artery (3). OCAD is a complex, slow and protracted process of evolution, closely related environment, diet, work and rest time, genetic factors with one or reciprocal action, inflammation invasion, metabolic disorders, myocardial cell remodeling, gradually infiltrate into the formation of coronary atherosclerosis, which affects oxygen supply to cardiomyocytes, hypoxia acceleration disease progression (4). Coronary artery stent implantation is commonly therapy of OCAD, but in recent years, there are more and more reports about restenosis after stenting. The academic literature on Boudoulas has revealed the appearance of several reverse subjects that coronary stent implantation or other interventional therapy accelerates the formation of coronary atherosclerosis in over a period of months to years, the course longer than for decades native disease taking shape on account of damage, repair, senility (5). For elderly patients, coronary stent implantation has a high risk, and many complications such as bleeding, hypercoagulability, thrombus shedding, electrolyte disturbance and other life-threatening conditions exist. Drug therapy still has some limitations. Thus, with the development of gene sequencing technology, it is necessary that helps to conduct novel strategies to treat OCAD by exploring the molecular mechanism from different perspectives.
In this research, we make a comprehensive analysis that comprise analyzing differentially expressed genes (DEGs) through the ratio of the diseased group to the normal group, functional gathering analysis incorporated gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG), drawing a protein interaction mapping, and filtration significant module and crux genes. Next, the multivariable logistic regression model was constructed between the clinical features and crux gene expression quantity, which can forecast the risk of OCAD. Finally, through the above analysis, we might have some fresh angle of view about the OCAD mechanism that may furnish increasing accurate guidance of diagnosis, treatment and prognosis.
Methods
Data acquisition
Import key phrase “obstructive coronary disease” in GEO database, looking for appropriate microarray data. In earnest searching, we obtain GSE90074 dataset by “GEOquery” package that about RNA sequencing in coronary atherosclerosis elderly, which include 50 non-OCAD samples and 93 OCAD samples as of 23 Jan 2019.
Analyzing DEGs
After obtaining the data, probe annotation was conducted according to the gene platform. It is common in a chip data that one gene might correspond to multiple probes, and multiple probes might correspond to one gene, due to base pairing principle and repeatability in a gene segment. Moreover, perform data cleaning to remove duplicate values and null values, testing data quality and standardized processing. At last, via the ratio of the diseased group to the normal group analyze DEGs by Limma package of R/Bioconductor project, calculating fold change, P value, correction P value and so on. The results of gene expression variation with a P value <0.01 were considered DEGs. We use mean plus two standard deviations of DEGs logFoldChange (logFC) to distinguish between up and down regulated genes, which have further advantages compared with directly using logFC multiple. In DEGs, greater than logFC_cutoff and zero is marked up regulated genes, less than −logFC_cutoff and zero marked down regulated genes, the rest of the genes labeled not.
logFC_cutoff = mean(|logFC|) + 2*sd(|logFC|)
Functional gathering analysis
The GO database contains ontology and GO annotation; ontology is deliberate the functions that interrelation with genes as the main body, GO annotation confirmed by published literature. Where genes are located in cells, what biological processes are involved, and how they function at the molecular level as the same with biological processes, cellular components, and molecular function, together explaining the functions of GO (6). KEGG database has extended drug and disease database, improved access system and expanded molecular object map to a higher level; for instance, genes, proteins, sugars and other molecular compounds reflect on diseases, drugs, modules, classifications and other pathways (7). The DAVID (Database for Annotation, Visualization, and Integrated Discovery) were executed functional gathering analysis incorporated GO and KEGG. P value <0.05 of the results was considered vital function in whole analysis.
Drawing a protein interaction mapping and module analysis
A string database (http://string-db.org) that gather and integrate largely information about the interrelationship between proteins, incorporate directly physical reciprocal actions and mediately functional reciprocal actions (8). It can help us more extensive realize the functions of genes and mutual relations. Import DEGs in string database draws a protein interaction mapping, and a combined score >0.4 was perceived as statistical meaning, using Cytoscape to beautify. Cytoscape explore biological networks with visualization and analyze molecular interactions, which provide highly interactive data sets and accommodate the plugin to expand the scope of function application, such as filtering module, function annotation, retrieval of literature (9). Afterwards, adding Molecular Complex Detection (MCODE) plugin in Cytoscape, identify significant module area from a protein interaction mapping. The conditions for filtration were as below: MCODE scores >10, degree cut-off =2, node score cut-off =0.2, Max. depth =100 and k-score =2. Whereafter, for the significant module conduct KEGG and GO functional gathering analysis in DAVID, and P value <0.05 was deemed as momentous meaning. Finally, using the CytoHubba (version 0.1) plug-in of the Cytoscape filtrate crux genes and analysis these biological process by BiNGO (version 3.0.3) module.
Statistical analysis
Related with clinical features set up a multivariable logistic regression model to predict the risk of OCAD with the gender, weight, history of diseases, and crux gene expression quantity incorporated. The R packages was conducted to analysis the process. In the beginning, all variable factors were screened to select the best matching group by the least absolute shrinkage and selection operator (LASSO) method. A principal advantage of the LASSO method is that can reduce the deviation of parameter estimation, which based on the least square method, generate the optimal variable parameters from the high dimensional variable data (10). Later, choose the characteristics with nonzero coefficients in the LASSO regression as the variables of the model (11). After LASSO screening, clinical information variables and crux genes expression quantity were included in the model to predicting the risk of OCAD. These characteristics have their own odds ratio (OR), 95% confidence interval (CI), and P value.
Ability to produce calibration curves to qualitatively evaluate the accuracy of the OCAD nomogram. In order to quantitative evaluation the model, measures Harrell’s C-index. At last, we adopt internal verification method to verify the model, and draw decision curve for clinical application through calculating the net benefits under different threshold probabilities in patients.
Results
Analyzing DEGs
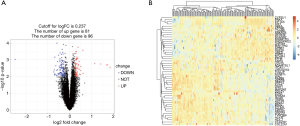
To distinguish the DEGs from the GSE90074 dataset that contains 50 non-OCAD patients and 93 OCAD patients in elderly crowd by using the Limma package (R project). The results of analysis generate totally 19,595 DEGs, after performing the filtration about the changed expression’s P value <0.01, 432 genes were considered the significant DEGs. Among them, incorporate 81 up- and 96 down-regulated genes. Figure1A presents all DEGs that red dots were marked 81 up-regulated DEGs, blue dots marked 96 down-regulated DEGs and the rest genes is black ones, the whole like volcanic eruption. The Figure1B shows heat-map that the expression amounts of top 50 genes in each sample include non-coronary artery disease and coronary artery disease patients and their similarity settlement relation.
Functional gathering analysis
To increase perception of obstructive coronary disease pathogenesis, comprehensive analyze 81 up- and 96 down-regulated DEGs by GO and KEGG pathway database. In GO biological processes, these DEGs’ functional gathering is mainly that positive regulation of endothelial cell apoptotic process, adjustment of immunoglobulin secretion and immunological response. For molecular functions, these DEGs closely related to endogenous or exogenous lipid antigen binding, jointing ubiquitin-protein ligase, linking zinc ion, and androgen receptor binding. In cellular component aspect, they are predominately located in the autophagosome membrane and clathrin-sculpted monoamine transport vesicle membrane. Moreover, the KEGG pathway enrichment analysis principally focus on the Toll-like receptor signaling pathway, Toxoplasmosis and Shigellosis. All results are detailedly illustrated in Table 1 and Figure 2.
Table 1
| Category | Term | Count | P value |
|---|---|---|---|
| GOTERM_BP_DIRECT | GO:2000353—positive regulation of endothelial cell apoptotic process | 3 | 0.005323547 |
| GOTERM_BP_DIRECT | GO:0050776—regulation of immune response | 6 | 0.013692822 |
| GOTERM_BP_DIRECT | GO:0051023—regulation of immunoglobulin secretion | 2 | 0.038984377 |
| GOTERM_CC_DIRECT | GO:0000421—autophagosome membrane | 3 | 0.014617802 |
| GOTERM_CC_DIRECT | GO:0070083—clathrin-sculpted monoamine transport vesicle membrane | 2 | 0.036498309 |
| GOTERM_MF_DIRECT | GO:0008270—zinc ion binding | 17 | 0.018911386 |
| GOTERM_MF_DIRECT | GO:0031625—ubiquitin protein ligase binding | 7 | 0.024546117 |
| GOTERM_MF_DIRECT | GO:0030883—endogenous lipid antigen binding | 2 | 0.038207923 |
| GOTERM_MF_DIRECT | GO:0030884—exogenous lipid antigen binding | 2 | 0.038207923 |
| GOTERM_MF_DIRECT | GO:0050681—androgen receptor binding | 3 | 0.038475526 |
| KEGG_PATHWAY | hsa04620: Toll-like receptor signaling pathway | 5 | 0.011254971 |
| KEGG_PATHWAY | hsa05145: toxoplasmosis | 5 | 0.012765579 |
| KEGG_PATHWAY | hsa05131: shigellosis | 4 | 0.015728246 |
BP, biological processes; CC, cellular components; DEGs, differentially expressed genes; GO, gene ontology; KEGG, Kyoto encyclopedia of genes and genomes; MF, molecular function.
Drawing a protein interaction mapping and module analysis
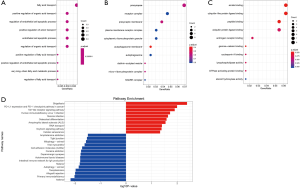
Through importing the 432 significant DEGs in string database, draws a protein interaction mapping that described straightforward relationship between proteins and predicted indirect relationships for them. Setting up the combined score more than 0.4, acquire a mapping that composed of 390 nodes and 853 edges, with the enrichment P value =7.07e−05. The mapping shows as Figure3A after beautify of Cytoscape. Next, using Cytoscape’s plug-in MCODE, screened the significant module from the mapping, which included 17 nodes and 107 edges. The Figure3B presents the network of significant module between protein and protein. Above them, the red is sign of up-regulation genes and the blue is sign of down-regulation genes. The significant module functions gathering analysis mainly use the tool of DAVID. Table 2 fully displayed the results; in GO database, this module is primarily situated on proteasome regulatory particle and membrane, influencing threonine-type endopeptidase activity and ubiquitin-dependent protein catabolic process. This module’s KEGG pathway concentrate mainly on proteasome and Epstein-Barr virus infection. Based on a protein interaction mapping, adopt 4 kinds of ranking method that is degree, radiality, MCC (maximal clique centrality), and EPC (edge percolated component) to calculate top 20 hub genes using CytoHubba plug-in. Whereafter taking the intersection, acquire 11 hub genes: BTRC, NFKBIA, PSMB1, PSMC1, COPS5, NEDD8, PSMA6, PSMD4, PSMD12, UBC, RPS4X. The details showed in Table 3. At the last, Figure3C displays that crux genes biological process using plug-in of BiNGO in Cytoscape.
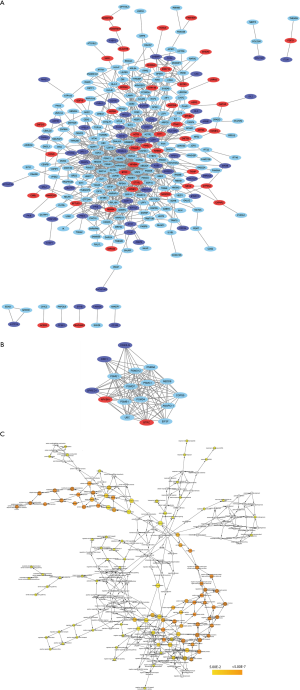
Table 2
| Category | Term | Count | P value |
|---|---|---|---|
| GOTERM_BP_DIRECT | GO:0006511—ubiquitin-dependent protein catabolic process | 2 | 0.09051555 |
| GOTERM_CC_DIRECT | GO:0005838—proteasome regulatory particle | 3 | 1.54E-05 |
| GOTERM_CC_DIRECT | GO:0016020—membrane | 4 | 0.047956647 |
| GOTERM_MF_DIRECT | GO:0004298—threonine-type endopeptidase activity | 2 | 0.021080797 |
| KEGG_PATHWAY | cjc03050: proteasome | 7 | 8.66E-11 |
| KEGG_PATHWAY | cjc05169: Epstein-Barr virus infection | 5 | 5.93E-05 |
Table 3
| Crux gene | MCC (score) | Degree (score) | EPC (score) | Radiality (score) |
|---|---|---|---|---|
| UBC | 8,082,716 | 51 | 97.062 | 8.019 |
| PSMA6 | 8,117,010 | 27 | 96.781 | 7.712 |
| NEDD8 | 7,997,095 | 33 | 96.628 | 7.821 |
| BTRC | 7,627,377 | 24 | 94.560 | 7.613 |
| PSMC1 | 8,116,080 | 21 | 94.791 | 7.654 |
| PSMD12 | 8,115,312 | 20 | 94.675 | 7.609 |
| PSMD4 | 8,106,648 | 19 | 94.589 | 7.637 |
| COPS5 | 726,889 | 21 | 94.357 | 7.695 |
| PSMB1 | 8,105,765 | 21 | 93.983 | 7.654 |
| RPS4X | 11,782 | 25 | 94.378 | 7.667 |
| NFKBIA | 3,669,890 | 21 | 93.493 | 7.773 |
Statistical analysis
According to 11 hub genes expression quantity, combining GSE90074 patients’ clinical information establish multivariable logistic regression model and draw nomogram to forecasting the risk of OCAD. The GSE90074 patients’ clinical information using “GEOquery” package extraction include gender, diabetes, hyperlipidemia, hypertension, BMI. As show in Table 4.
Table 4
| Clinical information | OCAD (n=93) | Non-OCAD (n=50) | Total (n=143) |
|---|---|---|---|
| Gender | |||
| Female | 36 | 32 | 68 |
| Male | 57 | 18 | 75 |
| BMI | |||
| 0 (<18.5 kg/m2) | 1 | 1 | 2 |
| 1 (18.5–25 kg/m2) | 22 | 10 | 32 |
| 2 (>25 kg/m2) | 70 | 39 | 109 |
| Diabetes | |||
| No | 55 | 33 | 88 |
| Yes | 38 | 17 | 55 |
| Hyperlipid | |||
| No | 19 | 22 | 41 |
| Yes | 74 | 28 | 102 |
| Hypertension | |||
| No | 11 | 6 | 17 |
| Yes | 82 | 44 | 126 |
OCAD, obstructive coronary artery disease.
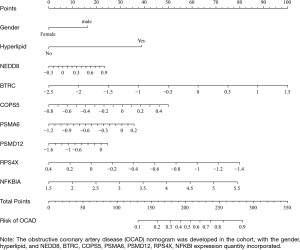
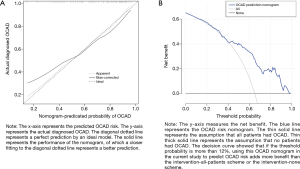
First of all, 16 kinds features decrease to 9 features after filtering with nonzero coefficients in the LASSO regression model, only having gender, hyperlipidemia, and NEDD8, BTRC, COPS5, PSMA6, PSMD12, RPS4X, NFKBIA expression quantity potential predictors. The filtration process is set out in Figure 4A,B. The logistic regression analysis’s coefficients, OR, 95% CI, and P value are presented in Table 5. The multivariable logistic regression model was exhibited as nomogram in Figure 5, which include above potential predictors, points of their correspondence and the risk probability of OCAD. Next, in Figure 6A, it is obviously that the calibration curve of nomogram improved that the multivariable logistic regression model has a good predictive power. The C-index of the model was calculated 0.786 (95% CI: 0.712–0.860), after internal random validation of setting up the bootstrapping =1,000, which was 0.731. In a word, the model was demonstrated that it can provide favorable discrimination for the risk of OCAD through the quantitative analysis of C-index. Finally, the Figure 6B indicated that the analysis of decision curve. For enhancing clinical applications, draws the decision curve, which displays that if the threshold probability is more than 12%, the model has extraordinary significance compared with no using it.
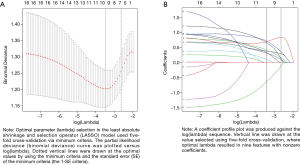
Table 5
| Intercept and variable | Prediction model | ||
|---|---|---|---|
| β | Odds ratio (95% CI) | P value | |
| Intercept | −1.939 | 0.144 (0.001–14.122) | 0.420 |
| Gender (male) | 0.461 | 1.586 (0.367–6.661) | 0.530 |
| Hyperlipid (yes) | 1.113 | 3.044 (1.289–7.397) | 0.012 |
| NEDD8 | 0.557 | 1.745 (0.130–23.186) | 0.671 |
| BTRC | 0.717 | 2.048 (1.137–3.793) | 0.019 |
| COPS5 | 1.103 | 3.014 (0.074–150.906) | 0.566 |
| PSMA6 | 0.731 | 2.077 (0.221–21.841) | 0.529 |
| PSMD12 | 0.392 | 1.480 (0.115–20.101) | 0.764 |
| RPS4X | −1.273 | 0.280 (0.023–2.880) | 0.300 |
| NFKBIA | 0.566 | 1.760 (0.649–5.680) | 0.301 |
Notes: β is the regression coefficient. CI, confidence interval.
Discussion
In current, with GO and bioinformatics developing, precise localization, planning and treatment is getting more and more attention. From gene molecules to clinical phenotypes, the entire process of disease development is crucial for treatment decisions. Prior researches have shown that the pathological basis of OCAD is mainly coronary atherosclerosis, which is involved various complex mechanisms that about endothelium dysfunction, inflammatory reaction, microvascular dysfunction, abnormal coagulation, apoptosis of macrophages, new angiogenesis in plaques (12). In our study, first of all, filtrating 432 significant DEGs, then use logFC_cutoff to distinguish 81 up- and 96 down-regulated genes. The literature on Curran-Everett revealed that the standard deviation represents the discrete degree of sample observations to the mean, and reckon the variability of a single observations in ensemble sample (13). The GO enrichment functional analysis of up and down regulated DEGs showed that these DEGs mostly located in autophagosome membrane and clathrin-sculpted monoamine transport vesicle membrane, associating with various biological processes, such as forward direction control of endothelial cell apoptotic process, adjustment of immunoglobulin secretion and immunological response. The molecular function mainly reflected in endogenous or exogenous lipid antigen binding, jointing ubiquitin-protein ligase, linking zinc ion, and androgen receptor binding. In addition, the enriched KEGG pathways primarily involved in toll-like receptor signaling pathway, toxoplasmosis, shigellosis. Essentially, the research on Ren has emphasized that overexpression of miR-330 in acute coronary syndrome enhances blood vessel endothelium cell multiplication via the WNT sign pathways and significantly suppresses atherosclerotic plaques formation (14). Fan’s academic literature revealed that Dectin-1 expressed by macrophages plays a pathogenic role during cardiac muscle ischemia reperfusion injuries through inducing macrophage polarized toward the M1 phenotype and neutrophil infiltration and Dectin-1-dependent pathway in the heart, through which the immune system may influence myocardial ischemia reperfusion injuries (15). In Zhao et al. research indicated the mutual effect of Fcγ receptors and immunoglobulin G antibodies bound to antigen response induce the activation of neutrophils, which enhances the destabilization of atherosclerotic plaque (16). Kounis and Hahalis’s study stated that elevated levels of immunoglobulin E is related to instability of the atherosclerotic plate and ischemic heart disease, which affects the development of atherosclerosis, highlighting that it can regard as early sign of atherogenesis and its results to predict probability of cardiovascular events (17). In Müller et al. literature found that suppressed ADAMTS7 with its metalloproteinase domain linking catalytic zinc ion, which might intervene coronary artery disease formation through effecting protein depolymerization during platelet response (18). The academic literature on Wu doctor has demonstrated that LincRNA-p21 through linking to mouse double minute 2 (MDM2) that is an E3 ubiquitin-protein ligase effecting degradation and acetylation of p53, strengthen p53 transcriptional activity, which influence vascular smooth muscle cells propagation and taking shaping neointima in the atherosclerosis process (19). The study of Nording et al. suggested that the relevance between oxidized lipids and designated complement components might increase to the platelet-lipid interaction in atherogenesis development (20). Saltiki et al. found that if the degree of coronary atherosclerosis obstruction is over 50% in postmenopausal women patients, the androgen receptor commonly present shorter stretch of polyglutamine compared with normal or mild lesion (21). Doddapattar et al.’s research showed toll-like receptor 4 sign pathway takes part in the progress that cellular fibronectin which contains extra domain A mediated deterioration of atherosclerosis (22). Başaran et al. found that the pathogenesis of polyarteritis nodosa or other vasculitis syndromes might be toxoplasma infection (23). Senthong et al.’s research showed that trimethylamine-N-oxide (TMAO), which is a metabolite extracting from intestinal microorganism, could associate with the pathogenesis of coronary artery disease and increasing cardiovascular event probability (24). Shigellosis maybe increases the risk of coronary atherosclerosis by affecting gut microbes. The above argument shows that DEGs functional enrichment analysis closely related to the occurrence of coronary atherosclerosis development and changes.
Moreover, draws a protein interaction mapping about DEGs to show interactions or connections, recognition the most significant module, including BTRC, EIF3F, ANAPC11, COPS5, NEDD8, PSMA6, CD40LG, ORC1, SPRED1, NFKBIA, UBC, PSMB1, PSMD4, PSMD7, PSMC1, PSME1, PSMD12. Later, filter 11 hub genes through 4 kinds arithmetic of CytoHubba plug-in, such as BTRC, NFKBIA, PSMB1, PSMC1, COPS5, NEDD8, PSMA6, PSMD4, PSMD12, UBC, RPS4X. It is not hard to find that ten crux genes are contained in the most significant module. The most significant module functions analysis, primarily situate in proteasome regulatory particle and membrane, to participate in the process of ubiquitin-dependent regulation protein metabolism, which impact the molecular activity of threonine-type endopeptidase. The KEGG pathway enrichment principally is Epstein-Barr virus infection and proteasome. Sharma’s research revealed that the process of protein degradation in the ubiquitin system may affect neuronal nitric oxide synthase driving angiotensin II in chronic heart failure (25). Jiang’s report found that the correlation between cardiovascular complications and Epstein-Barr virus infection (26). The research from Morris et al. indicated that coding variant rs1051338 decreasing the activity of lysosomal acid lipase (LAL) protein might increase the occurrence rate of coronary artery disease (27). In general, obstructive coronary heart disease is related to vascular endothelial injury, vascular inflammation, immune dysfunction, the metabolic disturbance of glucose, protein and lipid. The significant module and crux genes participate in various biological processes to influence these factors.
Finally, according to holistic thinking, genetic changes are a form of disease progression and related to environment, lifestyle, age, gender and other factors, sometimes heredity only a part. We are closely related to clinical features to analysis the relationship between gene expression and OCAD risk. BTRC, RPS4X, NFKBIA, COPS5, PSMA6, PSMD12, NEDD8, hyperlipidemia, and gender was involved in multivariable logistic regression model to forecast the risk of OCAD. BTRC is especially important in the model and have significant statistics meaning. Hypertension is usually risk factor in cardiovascular disease, but don’t included in the model, the cause which might it isn’t enough to discrimination due to high morbidity rate in older patients or lacking sample number.
BTRC encodes about 40 amino acid motifs being part of F-box protein family, which could mediate CD4 catabolism and involve in the degradation of NFKBIA (nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha). The F-box protein is element of subunits of ubiquitin protein ligase, taking part in the process of ubiquitin phosphorylation. NFKBIA encodes numbers ankrin repeat domains protein that classified to NF-kappa-B inhibitor family, which restrain NF-kappa-B/REL complexes participating in inflammatory responses through interaction with REL dimers. COPS5 encodes a protein that belongs to subunits in the COP9 signal body, which could participate in degradation of cyclin-dependent kinase inhibitor CDKN1B/p27Kip1 and be a specific coactivator of JUN/AP1 transcription factors. The COP9 signal body has been proved that positive control interaction with SCF-type E3 ubiquitin ligases. NEDD8 is a high homologous ubiquitin-like protein, which could combine lipids or target proteins to adjust their stability, activity, subcellular location and macromolecule mutual effect (28,29). Through commanding attachment of NEDD8 (neddylation) in the cullin RING ligases (CRLs) to affect the proteasome degradation, might selectively control protein metabolism in vascular smooth muscle (30). PSMD12 encodes protein about proteasome 26s subunit and non-ATPase 12, which could crack peptides through non-lysosomal pathway in an ATP or ubiquitin dependent process. Küry et al.’s research found that PSMD12 as holder subunits has tremendous biological meaning, impacting neurodevelopmental process (31). PSMA6 encodes protein about proteasome 20S subunit, the functions as the same with PSMD12. Wang et al.’s meta-analysis indicated that PSMA6-8C/G polymorphism is linked to gradually increasing morbidity of coronary artery disease, but exist differences in different ethnic (32). RPS4X encodes ribosomal protein S4, which could catalyze protein synthesis. Surprisingly, the model found that the lower expression of RPS4X can get higher score of predict the risk of OCAD. In summary, the above genetic analysis shows that the ubiquitin proteasome system has extraordinary effect in the metabolism of cardiomyocytes about OCAD through the regulation, degradation, signal transmission, injury repair and inflammatory immunity of various proteins.
Conclusions
In order to further learn about OCAD mechanism, compared with gene expression between OCAD and non-OCAD elderly patients, we have been recognition crux genes and their functions. All the DEGs might involve in various pathways about the OCAD occurrence and development, and we found that BTRC, NFKBIA, COPS5, NEDD8, PSMD12, PSMA6, RPS4X could provide great significant prognostic value for OCAD, which might help us to prophylaxis and treatment it. Nevertheless, there is still a long way to explore, existing some limitations such as small sample sizes, lacking of adequate validation and the differences of locations, times, and environments.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jxym-20-43). The authors have no conflicts of interest to declare.
Ethical Statements: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The research has no any ethical issues involved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Mayala HA, Yan W, Jing H, et al. Clinical characteristics and biomarkers of coronary microvascular dysfunction and obstructive coronary artery disease. J Int Med Res 2019;47:6149-59. [Crossref] [PubMed]
- Pizzi C, Xhyheri B, Costa GM, et al. Nonobstructive Versus Obstructive Coronary Artery Disease in Acute Coronary Syndrome: A Meta-Analysis. J Am Heart Assoc 2016;5:e004185. [Crossref] [PubMed]
- Mei WY, Liu LJ, Xu Q, et al. Additional Value of Early Repolarization Pattern in Prediction of Obstructive Coronary Artery Disease as Assessed by Coronary Angiography. Int Heart J 2019;60:296-302. [Crossref] [PubMed]
- Boudoulas KD, Triposciadis F, Geleris P, et al. Coronary Atherosclerosis: Pathophysiologic Basis for Diagnosis and Management. Prog Cardiovasc Dis 2016;58:676-92. [Crossref] [PubMed]
- Yahagi K, Kolodgie FD, Otsuka F, et al. Pathophysiology of native coronary, vein graft, and in-stent atherosclerosis. Nat Rev Cardiol 2016;13:79-98. [Crossref] [PubMed]
- The Gene Ontology Consortium. Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res 2017;45:D331-8. [Crossref] [PubMed]
- Kanehisa M, Furumichi M, Tanabe M, et al. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 2017;45:D353-61. [Crossref] [PubMed]
- Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res 2017;45:D362-8. [Crossref] [PubMed]
- Su G, Morris JH, Demchak B, et al. Biological network exploration with Cytoscape 3. Curr Protoc Bioinformatics 2014;47:8.13.1‐8.13.24.
- Vasquez MM, Hu C, Roe DJ, et al. Measurement error correction in the least absolute shrinkage and selection operator model when validation data are available. Stat Methods Med Res 2019;28:670-80. [Crossref] [PubMed]
- Kidd AC, McGettrick M, Tsim S, et al. Survival prediction in mesothelioma using a scalable Lasso regression model: instructions for use and initial performance using clinical predictors. BMJ Open Respir Res 2018;5:e000240. [Crossref] [PubMed]
- Koganti S, Eleftheriou D, Brogan PA, et al. Microparticles and their role in coronary artery disease. Int J Cardiol 2017;230:339-45. [Crossref] [PubMed]
- Curran-Everett D. Explorations in statistics: standard deviations and standard errors. Adv Physiol Educ 2008;32:203-8. [Crossref] [PubMed]
- Ren J, Ma R, Zhang ZB, et al. Effects of microRNA-330 on vulnerable atherosclerotic plaques formation and vascular endothelial cell proliferation through the WNT signaling pathway in acute coronary syndrome. J Cell Biochem 2018;119:4514-27. [Crossref] [PubMed]
- Fan Q, Tao R, Zhang H, et al. Dectin-1 Contributes to Myocardial Ischemia/Reperfusion Injury by Regulating Macrophage Polarization and Neutrophil Infiltration. Circulation 2019;139:663-78. [Crossref] [PubMed]
- Zhao N, Mi L, Zhang Y, et al. Altered human neutrophil FcγRI and FcγRIII but not FcγRII expression is associated with the acute coronary event in patients with coronary artery disease. Coron Artery Dis 2017;28:63-9. [Crossref] [PubMed]
- Kounis NG, Hahalis G. Serum IgE levels in coronary artery disease. Atherosclerosis 2016;251:498-500. [Crossref] [PubMed]
- Müller M, Kessler T, Schunkert H, et al. Classification of ADAMTS binding sites: The first step toward selective ADAMTS7 inhibitors. Biochem Biophys Res Commun 2016;471:380-5. [Crossref] [PubMed]
- Wu G, Cai J, Han Y, et al. LincRNA-p21 regulates neointima formation, vascular smooth muscle cell proliferation, apoptosis, and atherosclerosis by enhancing p53 activity. Circulation 2014;130:1452-65. Erratum in: Circulation 2019 Apr 23;139(17):e887. doi: 10.1161/CIR.0000000000000690. [Crossref] [PubMed]
- Nording H, Giesser A, Patzelt J, et al. Platelet bound oxLDL shows an inverse correlation with plasma anaphylatoxin C5a in patients with coronary artery disease. Platelets 2016;27:593-7. [Crossref] [PubMed]
- Saltiki K, Cimponeriu A, Garofalaki M, et al. Severity of coronary artery disease in postmenopausal women: association with the androgen receptor gene (CAG)n repeat polymorphism. Menopause 2011;18:1225-31. [Crossref] [PubMed]
- Doddapattar P, Gandhi C, Prakash P, et al. Fibronectin Splicing Variants Containing Extra Domain A Promote Atherosclerosis in Mice Through Toll-Like Receptor 4. Arterioscler Thromb Vasc Biol 2015;35:2391-400. [Crossref] [PubMed]
- Başaran Ö, Çakar N, Gür G, et al. Juvenile polyarteritis nodosa associated with toxoplasmosis presenting as Kawasaki disease. Pediatr Int 2014;56:262-4. [Crossref] [PubMed]
- Senthong V, Wang Z, Li XS, et al. Intestinal Microbiota-Generated Metabolite Trimethylamine-N-Oxide and 5-Year Mortality Risk in Stable Coronary Artery Disease: The Contributory Role of Intestinal Microbiota in a COURAGE-Like Patient Cohort. J Am Heart Assoc 2016;5:e002816. [Crossref] [PubMed]
- Sharma NM, Llewellyn TL, Zheng H, et al. Angiotensin II-mediated posttranslational modification of nNOS in the PVN of rats with CHF: role for PIN. Am J Physiol Heart Circ Physiol 2013;305:H843-55. [Crossref] [PubMed]
- Jiang S, Li X, Cao J, et al. Early diagnosis and follow-up of chronic active Epstein-Barr-virus-associated cardiovascular complications with cardiovascular magnetic resonance imaging: A case report. Medicine (Baltimore) 2016;95:e4384. [Crossref] [PubMed]
- Morris GE, Braund PS, Moore JS, et al. Coronary Artery Disease-Associated LIPA Coding Variant rs1051338 Reduces Lysosomal Acid Lipase Levels and Activity in Lysosomes. Arterioscler Thromb Vasc Biol 2017;37:1050-7. [Crossref] [PubMed]
- Santonico E, Nepravishta R, Mandaliti W, et al. CUBAN, a Case Study of Selective Binding: Structural Details of the Discrimination between Ubiquitin and NEDD8. Int J Mol Sci 2019;20:1185. [Crossref] [PubMed]
- Cappadocia L, Lima CD. Ubiquitin-like Protein Conjugation: Structures, Chemistry, and Mechanism. Chem Rev 2018;118:889-918. [Crossref] [PubMed]
- Martin DS, Wang X. The COP9 signalosome and vascular function: intriguing possibilities?. Am J Cardiovasc Dis 2015;5:33-52. [PubMed]
- Küry S, Besnard T, Ebstein F, et al. De Novo Disruption of the Proteasome Regulatory Subunit PSMD12 Causes a Syndromic Neurodevelopmental Disorder. Am J Hum Genet 2017;100:689. [Crossref] [PubMed]
- Wang H, Jiang M, Zhu H, et al. Quantitative assessment of the influence of PSMA6 variant (rs1048990) on coronary artery disease risk. Mol Biol Rep 2013;40:1035-41. [Crossref] [PubMed]
Cite this article as: Li J, Fan G. Identification of key genes and pathways in obstructive coronary artery disease by integrated analysis. J Xiangya Med 2020;5:14.